[All text is normative unless otherwise labeled]
A specification for content classification and content
interoperability in the clinical trial domain.
The key words “MUST”, “MUST NOT”, “REQUIRED”, “SHALL”,
“SHALL NOT”, “SHOULD”, “SHOULD NOT”, “RECOMMENDED”, “MAY”, and “OPTIONAL” in
this document are to be interpreted as described in [RFC2119].
[RFC2119] Bradner,
S., “Key words for use in RFCs to Indicate Requirement Levels”, BCP 14, RFC
2119, March 1997. http://www.ietf.org/rfc/rfc2119.txt.
[1] Media Types. IANA. [Online] http://www.iana.org/assignments/media-types/media-types.xhtml
[2] OWL 2 Web
Ontology Language, Structural Specification and Functional Style Syntax,
W3C Recommendation. W3C OWL Specifications. [Online] October 27, 2009.
http://www.w3.org/TR/owl2-syntax/#Data_Properties
http://www.w3.org/TR/owl2-syntax/#Annotation_Properties
1.3 Non-Normative
References
[3] NCI EVS. National Cancer Institute. [Online] http://evs.nci.nih.gov/.
[4] Dublin Core Metadata. [Online] http://dublincore.org/.
[5] Enabling Tailored Therapeutics with Linked Data. Anja
Jentzsch, Oktie Hassan Zadeh, Christian Bizer, Bo Andersson, and Susie
Stephens. s.l. : In Proceedings of the WWW2009 Workshop on Lined Data on the
Web, 2009.
[6] Clement Jonquet, Paea LePendu, Sean Falconer, Adrien
Coulet, Natalya Noy, Mark Musen, and Nigam Shah. Ontology-based Search and
Mining of Biomedical Resources. Semantic Web Challenge 2010 Submission.
[Online] 2010. http://www.cs.vu.nl/~pmika/swc/submissions/swc2010_submission_4.pdf,
2010..
[7] The Protégé Ontology Editor and Knowledge Acquisition
System, Protégé ontology editing tools and open source community. [Online]
Stanford Center for Biomedical Informatics Research(BMIR), Stanford University.
[Cited: February http://protege.stanford.edu,
2012.]
[8] Object Management Group, Business Process Model and
Notation (BPMN) Version 2.0, OMG. [Online] January 3, 2011. http://www.omg.org/spec/BPMN/2.0/.
[9] A Comparison of Semantic Markup Languages. Varun
Ratnakar, Yolanda Gil. Pensacola, Florida : In proceedings of the 15th
International FLAIRS Conference, Special Track on Semantic Web, 2002.
[10] Semantic Infrastructure to Enable Collaboration in
Ontology Development. P.R.Alexander, C.I.Nyulas, T.Tudorache, T.Whetzel,
N.F.Noy, and M.A.Musen. Philadelphia : In Proceedings of The International
Workshop on Semantic Technologies for Information-Integrated Collaboration
(STIIC 2011), 2011.
[11] NCBO and BioOntology BioPortal. [Online] National
Center for Biomedical Ontology (NCBO), February 2012. http://www.bioontology.org/.
[12] NCI Thesaurus Online Vocabulary. [Online] National
Cancer Institute (NCI), February 2012. http://ncit.nci.nih.gov/
[13] CarLex Content Models for Health Science. NCBO
BioPortal. [Online] http://bioportal.bioontology.org/ontologies/3008/?p=terms
Many organizations in the health sciences industry –
BioPharma and Healthcare – use Enterprise Document Management Systems (EDMS) to
manage and archive clinical trial documents and records. Although many
organizations coordinate and share the same documents, organizations lack a
standards-based metadata vocabulary and method to classify and share electronic
clinical trial documents, electronic medical images and related records.
Additionally, it is difficult to efficiently search, report, and audit sets of
clinical trial documents and their associated records due to a lack of a common
metadata vocabulary. For example, if an organization wishes to search for a set
of documents from the country ‘France’, unless each document is tagged with the
metadata term ‘Country’, it would be very difficult to find such documents
among distributed sets of clinical trial data. Information is often difficult
to locate, unless it is indexed with a common published set of metadata
vocabulary terms. This lack of interoperability among digital content
repository resources, due to vocabulary and schema differences, makes rapid
secure information discovery, retrieval, exchange, and sharing difficult for
organizations.
Central to our vision is the belief that organizations that
create document repositories should have the flexibility to classify, name, and
organize documents in a way that meets their business needs and yet have
interoperability, i.e., the ability to rapidly search and share repository
resource information with other organizations in a standard format that is
based on open systems standards.
As clinical trial stakeholder organizations seek to move
from paper-based record-keeping to electronic approaches, information
interoperability, information standards and agency compliance are key factors
in accelerating the safe delivery of therapies to patients.
In order to move clinical trial content from paper-based
approaches to automated electronic Document Management Systems in the cloud,
on-premises (in network) or offline, a standardized machine readable content
classification system, with a web standards-based controlled metadata vocabulary,
is needed. For those with access to Electronic Document Management Systems
(EDMSs), a method to exchange content between systems is needed. For those
without access to EDMSs, a method to exchange, view, and navigate content
offline is needed. Ideally anyone with a web browser and proper permissions
should be able to view the records and documents exported from an EDMS.
In the clinical trial domain, documents, medical images, and
other electronic content are typically stored in an electronic archive known as
the electronic Trial Master File (eTMF). The eTMF serves as a central
repository to store and manage essential clinical trial documents and content
as well as preparing content for regulatory submissions. Today, there is no
standard that defines how eTMF documents and records should be formatted for
electronic export and exchange between systems. To maximize interoperability,
it is important to adopt an open systems approach that is standards-based,
operating system independent, software application independent, and computer
language independent.
Finally, any eTMF system must support government agency
requirements for exported electronic records. The use of a standards-based,
agency supported electronic document export formats will help raise the
effectiveness, efficiency and safety of clinical trials and will help
organizations share higher quality information more efficiently.
The purpose of the OASIS eTMF Standard Specification is to
define machine readable formats for clinical trial electronic Trial Master File
(eTMF) content interoperability and data exchange, a metadata vocabulary, and a
classification system that has a set of defined policies and rules. This goal
is achieved by specifying:
a) An eTMF
content classification model, which is comprised of a standards-based metadata
vocabulary and a content classification ontology;
b) A set of eTMF
content classification rules and policies;
c) An eTMF
Data Model.
Features supported in the OASIS eTMF Standard Specification
are divided into the following categories:
1. Core
Technology Architecture
2. Content
Classification System
3. Core
Metadata and Content Type Term Sources
4. Content
Model
5. Data Model
6. eTMF
Metadata Vocabulary for Content Classification
7. eTMF
Metadata Vocabulary for Content Tagging
Each of these categories is discussed in a separate section.
The benefits of implementing interoperable systems that
enable data sharing among clinical trial stakeholders are obvious and evident.
Everyone who has used the internet experiences the benefits of interoperability
– the ability to open and view a web page in a browser is premised on
interoperability capabilities and web standards. Regardless of who authored or
who hosted the content, users are able to view the content in a web browser.
The high level benefits of a standard for interoperable
clinical trial information exchange that is based on web standards are
summarized below:
·
Accelerate
clinical trial development timelines with interoperable data exchange
·
Streamline
agency compliance with standards-based exports and eSubmissions
·
Enable
sharing between clinical trial stakeholders with language independent
taxonomies
·
Enhance
clinical trial safety and efficacy with serious adverse event data exchange
The key OASIS eTMF foundational layers, as illustrated in Figure 1, include a Content Classification System (CCS) layer to automate content
classification; a Vocabulary for Content Management Layer to describe
classifications and documents through published vocabulary; and a Web Standard
Technology Core Layer, which includes W3C standards for information discovery
and exchange in addition to support for electronic and digital signatures and
business process models that reduce paper handling processes.
Figure 1: eTMF Foundational Layers
|
|
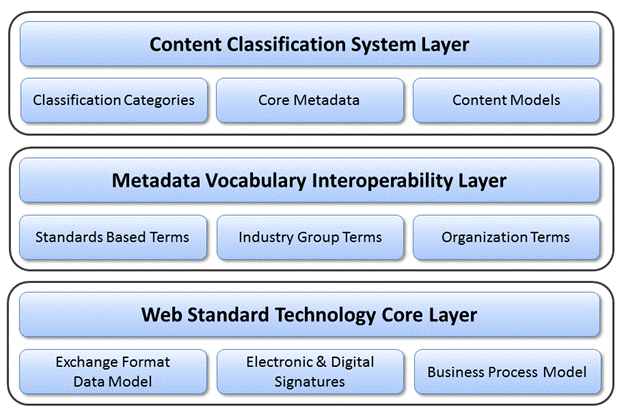
The first layer is the Content Classification System
(CCS) layer, which includes three components: Classification Categories
Component, Core Metadata component, and Content Model (Taxonomy) component.
Details of this layer are discussed in Section 5, “Content Classification System” on page 11.
The second layer is the Metadata Vocabulary
Interoperability Layer. The OASIS eTMF model utilizes a controlled vocabulary
for content management that is based on terms curated by the National Cancer
Institute’s NCI Thesaurus Enterprise Vocabulary Services (NCI EVS) [3]. NCI
EVS’ term database is used globally, and contains terms used by healthcare and
life sciences standards organizations such as HL7, CDISC, FDA, NIH and others.
As part of its term curation effort, NCI manages the semantic relationships
between terms and publishes its term database in a machine readable format
known as RDF/XML; a web standard that enables interoperable data exchange
between systems. Just as the internet uses the HTML to exchange data on
websites, the OASIS eTMF Standard’s metadata vocabulary layer is based on the
RDF/XML web standard so that any language or term names can be used in the
presentation layer. Data interoperability is maintained through the use of
RDF/XML in the metadata vocabulary layer, and is separate from the term label that
end users see.
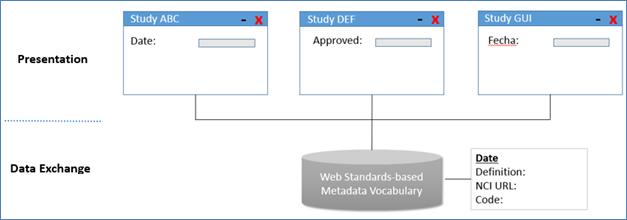
Figure 2: Presentation flexibility and data exchange
interoperability with web standards-based metadata vocabulary
In addition to providing organizations with the ability to
localize and customize any term label through the use of the Display Name
metadata attribute, the metadata vocabulary layer provides organizations with
the ability to create cross-referenced taxonomies that have a common
interoperable data. As an example, a sponsor and a CRO organization might use
a different term to represent the same content in an eTMF. The use of display
names enables these organizations to share data seamlessly regardless of the
names or even language used to display the terms.
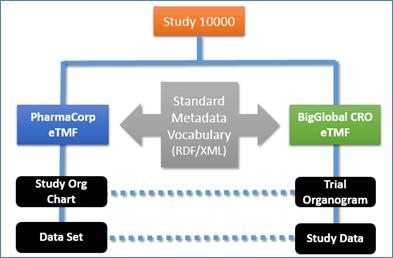
Figure 3: Multiple study taxonomies can be used with different names yet the same data.
The metadata vocabulary layer is flexible and can be
extended using a set of metadata vocabulary policies. The metadata vocabulary
layer contains standards-based terms, terms sourced from industry groups, and
organization-specific internal terms. The OASIS eTMF model metadata vocabulary
layer defines a standard set of metadata vocabulary terms that are present in
all OASIS eTMF Standard content models, enabling interoperable exchange of
taxonomies between sponsors, contract research organizations, investigators and
other stakeholders in the clinical trial ecosystem. Support for the DIA TMF
Reference Model (TMF RM) is provided in the OASIS eTMF Model through a
cross-reference mapping of terms. The DIA TMF RM provides a set of terms for
the industry through its published spreadsheet. The OASIS eTMF Model provides a
cross-reference mapping of the NCI-based OASIS metadata vocabulary terms to DIA
TMF RM terms, providing a path forward to a global ISO standard under the OASIS
eTMF Standards effort.
The third layer is the Web Standard Technology Core
layer. This layer is based upon the W3C RDF/XML, which represents resources
through the web, can be easily searchable, widely used, and allows sharing
electronic information through the cloud.
The CCS Layer includes Classification Categorization
(through a classification hierarchy or Taxonomy), a Metadata component, which
characterizes content, and a Content Model component, which includes a
published set of classification metadata for a specific domain (e.g., eTMF).
These components are described in the following sub-sections.
Similar to
how the Dewey Decimal system is used to classify books by category in a
library, the OASIS eTMF model classification categories component utilizes
Categories and Content Types with a decimal numbering format to classify
documents or content items. The classification categories component format is
based on the Universal Decimal Classification System (UDC),
which is widely adopted and used by libraries in over 170 countries worldwide.
This component is designed for both human and machine readability. It allows
for automated sorting of content classifications and documents and has a
flexible and infinitely expandable hierarchical system that can use any
vocabulary or text-based terms.
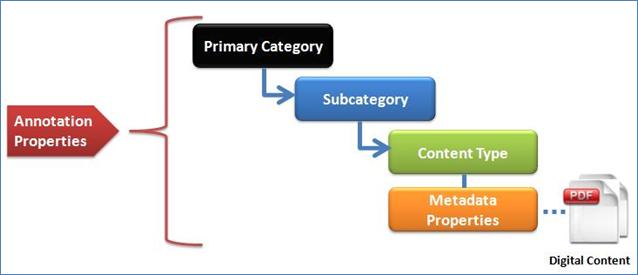
Figure 4: The OASIS eTMF Classification Categories Hierarch
The
classification categories component uses a hierarchy of classifications and a
numbering scheme to classify documents and content in unique categories. To
maximize machine readability, the classification and numbering scheme is based
on the W3C XML naming conventions. In this naming convention, only simple text
is allowed for category naming and numbering, and special characters, such as (
) *$ @ ! and others are prohibited. Classification numbers use a digital dot
notation where leading zeros are prohibited to conform to the W3C XML naming
conventions.
The
classification categories component contains classification entities, such as Categories,
Sub-Categories, and Content Types. Content Items, such as
documents or images, are linked to Content Types. Metadata Properties are
linked to Content Types to provide information about Content Items. Annotation
Properties provide information about Classification entities and Metadata
properties. An example for the Classification Categories hierarchy is shown in Error! Bookmark not defined., where a document or digital content is
classified in Categories. Each digital Content Item is classified by a Primary
Category, linked to a Sub-Category, with a link to a single specific Content
Type. Annotation Properties (1) provide annotations for Categories,
Sub-Categories, and Content Types. As an example, the Annotation Property
‘Definition’ describes briefly the meaning of the title assigned to a Category,
a Sub-Category, or a Content Type. The W3C OWL2 specification provides a general
description of Annotation Properties. Further details about Annotation
Properties are provided in the Appendix.
The Primary
Categories in the eTMF domain are numbered from 100 – 199, providing 100
primary category divisions. Use of the three digit primary category yet
provides additional categories for future growth in other health science domain
areas such as legal, administrative, research and development or other domain
areas, from 200-999. An example of how content might be classified, named, and
numbered using the OASIS eTMF content classification model is shown below.
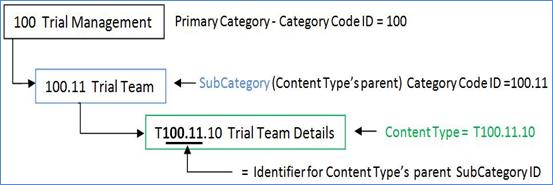
Figure 4: OASIS eTMF
Classification Category Code Numbering
Classification categories
terms are uniquely numbered using a hierarchical numbering scheme with a
digital dot notation. The classification category numbers, known as Category
Codes, allow content to be automatically classified in the content tree
hierarchy, as shown in Figure 4. Each classification number forms a unique
identifier that describes the position of the content in the hierarchy, making
documents or content items easily sorted and searched. Design and naming of
classification categories in addition to rules related to Category creation and
modification are all discussed in the following section.
Every
Category, Sub-Category, and Content Type contains Annotation Properties.
Content Types have both annotation properties and metadata assigned to them, as
illustrated in Figure 5. Each Classification entity contains at least the
following annotation properties: Category Name, Category Code, and Term Type.
Further details about Annotation Properties are provided in the Appendix.

Figure 5: Classification Hierarchy Relationships,
Annotation Properties, and Metadata
Primary Categories and Sub-Categories
are assigned unique decimal numbers for identification and classification called
Category Code numbers. They correspond to one and only one Category
Name. Content Types are linked to a parent Category and are used to link
related documents or content items. Additionally, the Category Code for any
Content Type has a unique numeric code prefixed by the letter “T” for
identification and classification. The Category Code for a Content Type is
known as Content Type ID. A Content Type ID always contains its parent
Sub-Category’s Category Code number in the numeric prefix of the Content Type
ID, as illustrated in Figure 6.
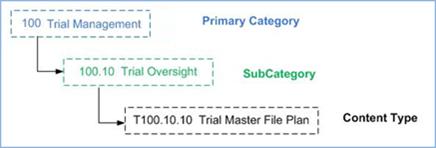
Figure 6: Classification
hierarchy example showing categories and classification hierarchy names
The Classification System follows a
naming scheme that combines the classification hierarchy name (i.e., Category
Code, which is designed to automate document classification and locate the
category in the content model hierarchy) and the simple text-based name (i.e.,
Category Name, which is used in the OASIS eTMF model for compliance with other
standards). An example is the name “100.10 Trial Management” assigned to a
Primary Category, where the first part “100.10” reflects the Category Code,
while the second part “Trial Management” represents the Category Name.
Generally, all text-based names, or
Category Names, should be unique and should not start with a number or a
special character to be compatible with XML naming standards. In the following,
we summarize the scheme used in generating the classification hierarchy name:
·
Primary Categories: Have a 3-digit numbers starting from
100: 100, 101, 102, etc. up to 999. The maximum number of Primary Categories is
900. Primary Categories act as headings, which include Sub-Categories. Primary
Categories may have zero or more Sub-Categories as their children.
·
Sub-Category: The Sub-Category classification hierarchy
number is based on the number assigned to the Sub-Category’s parent category.
It is a sequence number delimited with a period, to indicate the Sub-Category
division. Numbers for sub-categories start from 10: 10, 11, 12, etc., up to
99. The maximum number of nested sub-category items per parent category is 90.
Examples are: 100.10, 100.11, 100.12, and 142.23.67. The maximum number of
Sub-Category divisions is 5 excluding the 3-digits for the Primary Category
(i.e., aaa.bb.cc.dd.ee.ff). Every Sub-Category must have one and only one
Primary Category or Sub-Category as its parent. Sub-Categories may have zero or
more Content Types as their children.
·
Content Type: This is a two digit number sequenced from
10-99. It is based on the Content Type’s parent Sub-Category and a sequence
number delimited with a period to indicate the Content Type ID, as illustrated
in Figure 7. The Content Type ID is always prefixed with the letter “T” to
indicate that it is a Content Type entity. Content Types have one and only one
Sub-Category as their parent. Typically, a Content Type ID number includes its
hierarchical position in the Content Classification hierarchy.
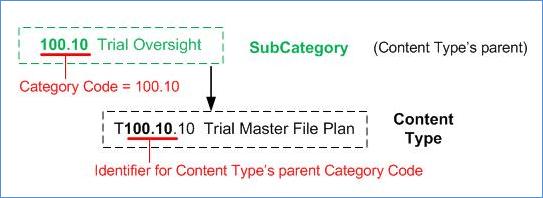
Figure 7: Content Type ID
Structure
An implementation example for a content repository is shown
in Figure 8. Within a repository, a single archive would typically contain a
collection of related content, such as those relevant to a specific clinical
trial study. This figure shows the archive of a clinical study labeled “Study
102880.” Examples for Category numbering (e.g., “100 Trial Management
Category”), Sub-Category numbering (e.g., “100.10 Trial Oversight
Subcategory”), and Content Type numbering (e.g., “T100.10.10 Trial Master File
Plan”) are illustrated in the figure in different colors.

Figure 8: Content
Classification Scheme for a Clinical Trial Example
Often organizations use abbreviated names, organization
specific names, or localized language names for some or all of the
Classification Entities. The eTMF model allows applications to use an
abbreviation as the Classification Entity Name and use an optional internal
label. For interoperability, when using an abbreviation or internal name with
any Entity, make sure to retain the original Code for Categories and Content
Types.
In an OASIS eTMF model implementation, a new Category,
Sub-Category, or Content Type could be added to the classification hierarchy,
as long as the classification numbering format is followed and the new entity
does not conflict with any existing item. Addition of Categories and Content
Types has two possibilities: the new Category or Content Type can either be Organization-specific
or Domain-specific.
In the first case, the details of the Organization-specific
Category or Content Type are entered by the user and a Category Code and a
machine-readable unique Term Code are generated locally in a way that ensures
the classification numbering format is followed and no conflicts exist in the
classification hierarchy. Category Codes and Term Codes for internal use can be
generated following the published Category Code format. These codes should have
the ‘Z’ prefix as illustrated in Table 1. Note that, Organization-specific
Classification Categories and Content Types could also be imported to a content
model (e.g., one organization publishes its own Classification Categories and
Content Types for public use and another organization uses them). However, the
importing party must check for conflicts, as Organization-specific
Classification Categories and Content Types are not interoperable.
In the second case, the Domain-specific Category or
Content Type is imported (through looking up the term) from a published Content
Model. In this case, the Category Code could be modified to enable placing the
new Category in the classification hierarchy. However, the assigned Code should
not be changed under any circumstances, so as not to violate interoperability.
Generally, new Categories, Sub-Categories, and Content Types could only be
imported from existing Content Model Categories, Sub-Categories, and Content
Types, respectively ( i.e., no mixing is allowed, such as importing an existing
Content Model’s Content Type as a new Sub-Category).
Restrictions apply to which Annotation Properties can be
modified for different types of Classification Categories and Content Types.
Except for annotation properties of Organization-specific classification
Categories and Content Types, only Display Name, Definition, Abbreviation, and
Requirement Annotation Properties can be modified. Table 1 illustrates rules for modifying/creating annotation properties for different types of
Classification Categories and Content Types. Please refer to the Appendix for
detailed information regarding Annotation Properties. It is worth mentioning
that domain specific classifications are considered to be the core for the content
model instance in use. Additionally, a content model could use more than one
domain.
Table 1: Rules for Creating and
Editing Annotation Properties of Categories and Content Types
|
Type of Category or
Content Type Term/ Create or Edit Annotation Property
|
Display Name
|
Preferred Name
|
Requirement
|
Definition
|
ValueSet
|
Abbreviation
|
Data Type
|
Term Source URL
|
|
Domain-specific
|
Yes
|
No
|
Yes
|
Yes
|
No
|
Yes
|
No
|
Yes
|
|
Organization-specific
|
Yes
|
Yes (1)
|
Yes
|
Yes
|
Yes
|
Yes
|
Yes
|
No (2)
|
|
(1) Preferred Name can
be created but not edited.
(2)
Organization-specific
term has no Term Source URL.
|
Domain-specific Classification Categories and Content Types
cannot be deleted from the content model. Instead of deleting a classification
item that is unused, the item’s name is marked as ‘Reserved’, to denote that
the item is not used in the content model. This marking enables
interoperability and the future use of the item, if needed. However,
Organization-specific Categories and Content Types can be deleted from the
Content Model. Table 2 provides a summary of the OASIS eTMF modification rules
discussed in this section.
(Continued)
Table
2: Rules for Addition, Modification, and Delete of Classification Categories
and Content Types
|
Type of
Classification Category or Content Type/Action
|
Import Term
|
Generate Code
|
Add / Modify
Term
|
Delete/Reserve
Term
|
|
Domain-specific
|
Yes
|
No
|
Yes/Yes
|
Reserve/UnReserve
|
|
Organization-specific
|
Yes
|
Yes
|
Yes/Yes
|
Delete
|
Metadata is used to give additional information about
digital content items and classification categories. The OASIS eTMF model
includes two types of metadata (illustrated in Figure 9):
·
Metadata
Properties: Describes the Content Type, e.g., Study ID, Site ID, Org, etc. It
is also called Data Properties.
·
Annotation
Properties: Describes attributes of Content Classifications and attributes of
Metadata Properties.
Each of these metadata types are discussed in detail in the
following sections.
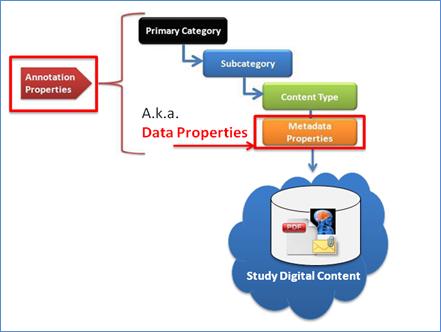
Figure 9: OASIS eTMF Metadata
Definitions
What is metadata and why is it needed? Metadata, or
information about data, is used to tag or index digital content items such as
documents. In the context of a Content Management System (CMS), metadata is
used to help organizations automate the classification and search, report, and
exchange of digital content items. Every digital document has some basic core
metadata. For example, every computer file contains metadata such as the ‘file
type.’ That metadata facilitates file exchange and enables applications to
automatically process files. Figure 11 shows an example for file metadata,
displayed upon right-clicking on it.

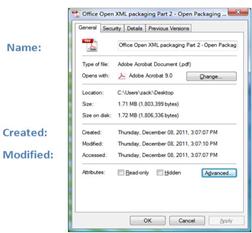
Figure
10: File Metadata Example
|
|
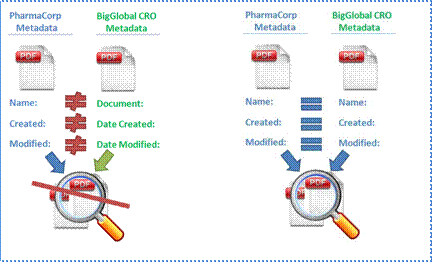
If every organization uses different metadata terms (as
shown in Figure 12), it is impossible to enable efficient global search,
reporting, and classification of documents within and outside of an
organization.
As illustrated in the figure, the use of dissimilar metadata
terms inhibits efficient search and decreases productivity, while the use of
similar terms enhances search efficiency and business productivity.
The OASIS eTMF model metadata is divided into four
types: Core Metadata, Domain Specific Metadata, General
Metadata, and Organization Specific Metadata.
·
Core
Metadata is associated with every document or Content Item, such as the basic
audit information ‘Created by’ and ‘Modified by’, which are widely used in
EDMSs.
·
Domain-based
Metadata are those used in specific domain areas, such as the metadata used in
the Clinical Trials electronic Trial Master File (eTMF) domain. General
Metadata is a set of Metadata Properties obtained from public terminology
resources and metadata vocabularies (in support of interoperable solutions),
such as Dublin Core Metadata [4].
·
Finally,
in order to allow for a flexible model that organizations can easily adapt to
existing business terms and business processes, Organization Specific metadata
can be used with any content type. In general, Organization specific metadata
terms, unless widely published, may not be interoperable with content models in
use by other organizations or entities. Due to its lack of interoperability,
Organization specific metadata is not published in the OASIS eTMF model.
Domain-specific, Organization-specific, and General Metadata Properties could
be added to a Content Model according to an organization’s needs. Please refer
to the Appendix for further details.
|
|
|
|
|
|
 |
|
|
Figure 11: Similar vs. Dissimilar Metadata or Terms
|
|
The OASIS eTMF model metadata is
also known as Data Properties (both names are used interchangeably
throughout this document). Data Properties are similar to XML
attributes. They define the attributes of Content Types. For example, ‘Country’
is a Data Property, which describes the country for a Content Item. Data
properties are associated with each Content Type instance. Data Properties are
simple Content Item tags without explicit relationships defined within the
model. For a general description of a Data Property, see the W3C OWL2 specification [2]. Data
properties have a number of annotation properties that provide additional
information.
To ensure interoperability in the OASIS eTMF model, a number
of rules must be followed when adding, modifying, or deleting metadata terms,
as summarized in Table 3 and Table 4 . Adding Domain-specific Metadata terms,
General Metadata terms, and Organization-specific Metadata terms is allowed.
Metadata terms could be added to the content model through looking up (i.e., importing)
or enabling the user to enter its details, as shown in Table 3. The former case applies to General and Domain-specific Metadata terms (as their details are
already available in other published content models or in resources for public
vocabularies), while the second case applies to Organization-specific Metadata
terms. Core Metadata terms is part of the published content model and cannot be
added.
Generally, interoperability is satisfied through the Code
annotation property included with every entity term. This annotation property
provides a unique number assigned to every entity term, including Category,
Sub-Category, Content Type, and Metadata terms. To ensure interoperability,
this unique code value cannot be modified once created.
In case of Organization-specific metadata terms, the details
of these terms (i.e., values of annotation properties) are
user-defined with exception to Code annotation property, which is generated
locally. Domain-specific and General Metadata terms could be imported through
looking up existing sources, such as the NCI Thesaurus and Dublin core,
respectively. When importing metadata terms, the Code annotation property
should be retained unmodified. Note that, some General Metadata properties may
not include the Code annotation property. To ensure interoperability, these
codes should be generated locally whenever required. The local generation of
Codes should ensure the uniqueness of the generated code such that no conflicts
exist with any other codes in the content model. Rules for Code generation and
the import of different types of metadata terms are summarized in Table 3. This table also presents rules for addition and modification of metadata
terms (in the content model) according to their types.
|
Metadata Type/Action
|
Import Term
|
Generate Code
|
Add / Modify
Term
|
Delete/Reserve
Term
|
|
Core
|
No
|
No
|
No/Yes
|
No/No
|
|
Organization-specific
|
No
|
Yes
|
Yes/Yes
|
Delete
|
|
Domain-specific
|
Yes
|
No
|
Yes/Yes
|
Delete
|
|
General
|
Yes
|
Yes(1)
|
Yes/Yes
|
Delete
|
|
(1)
Only
generate Code if a code is not available from the term source (e.g., Dublin
Core metadata)
|
Table 3: Rules for Addition,
Modification, Import, and Delete of Metadata Properties
Modification of metadata terms is allowed; however, not all
annotation properties could be modified. Table 4 illustrates annotation
properties that could be modified per metadata type.
|
Metadata Type/ Create or
Edit
|
Display Name
|
Preferred Name
|
Requirement
|
Definition
|
ValueSet
|
Abbreviation
|
Data Type
|
Term Source URL
|
|
Core
|
Yes
|
No
|
No
|
Yes
|
No
|
Yes
|
No
|
No
|
|
Organization
Specific
|
Yes
|
Yes (1)
|
Yes
|
Yes
|
Yes
|
Yes
|
Yes
|
Yes
|
|
Domain
Specific
|
Yes
|
No
|
Yes
|
Yes
|
No
|
Yes
|
No
|
No
|
|
General
|
Yes
|
No
|
Yes
|
Yes
|
No
|
Yes
|
No
|
Yes (3)
|
|
(1) Preferred Name can
be created but not edited.
(2) Only allow editing
of a URL if there is no URL available from the term source.
|
Table 4: Rules to
Edit/Create Metadata Annotation Properties
Finally, when a published content model is used, some
metadata terms cannot be deleted; they are labeled ‘reserved’ in the content
model, as shown in Table 3. Whenever these terms are used in the future, they
would be labeled as ‘unreserved’ metadata terms. Domain-specific Metadata,
Organization-specific Metadata, and General Metadata can be deleted from the
content model if they are not referenced in any content item. Core Metadata
terms are not allowed to be reserved or deleted, as they provide basic
information in any content model.
Annotation properties provide additional information about
different entities in the OASIS eTMF model, including classification categories
(i.e., Categories and Sub-Categories), Content Types, and Metadata Properties.
An example for annotation properties is Code, which uniquely identifies
different terms in a content model. The OASIS eTMF model includes two types of
annotation properties: Core and Organization-specific. The former
are part of the published content model, while the latter are added to the
content model according to an organization’s specific needs.
As with Classification Categories, Content Types, and
Metadata Properties, Annotation Properties also have rules for modification,
addition, and deletion. Table 5 summarizes rules for addition, deletion,
modification, and import of annotation properties. Generally, Core annotation
properties are part of the published content model and they can be neither
added nor deleted. Both core and organization specific annotation properties
cannot be imported (i.e., looked up in public resources). Additionally, when an
organization-specific annotation property is added, a unique code is generated
locally. This code should not conflict with any other existing code.
|
Annotation Properties
Type/Action
|
Import Term
|
Generate Code
|
Add / Modify Term
|
Delete/Reserve Term
|
|
Core
|
No
|
No
|
No/Yes
|
No delete or
reserve
|
|
Organization-specific
|
No
|
Yes
|
Yes/Yes
|
Delete
|
Table 5: Rules for Addition,
Modification, Import, and Delete of Annotation Properties
Annotation properties also have additional annotation
properties that provide details about them. In case of modifying details of
annotation properties, only a subset of the annotation properties of annotation
properties could be modified for the Core type, while more annotation
properties could be modified for the Organization-specific type. Annotation
properties that can be created or modified are illustrated in Table 6.
|
Annotation Properties
Type/ Create or Edit
|
Display Name
|
Preferred Name
|
Requirement
|
Definition
|
ValueSet
|
Abbreviation
|
Data Type
|
Term Source URL
|
|
Core
|
Yes
|
No
|
Yes
|
Yes
|
No
|
Yes
|
No
|
No
|
|
Organization-specific
|
Yes
|
Yes (1)
|
Yes
|
Yes
|
Yes
|
Yes
|
Yes
|
Yes
|
|
(1) Preferred Name can
be created but not edited.
|
Table 6: Rules to
Edit/Create Annotation Properties of Annotation Properties
Finally, the details of the OASIS eTMF model annotation
properties are presented in the Appendix.
Content models represent content classifications,
relationships, and metadata in a semantic web taxonomy or ‘Ontology’. Content
models are technology agnostic; there is no particular software, computer
operating system, or application required in order to use them. They are
published using a Web standard format known as ‘OWL’. This format allows
browser-based discovery of information. To illustrate how the OASIS eTMF
content model can be used in an organization to manage content items, consider
the technology that nearly everyone has used - management and organization of
music MP3 files.
All MP3 files today contain standard tags or metadata, such
as Artist, Title, Album, and Genre that are embedded in the MP3 file to enable
rapid electronic classification and search. These tags describe the music or
album. A user typically classifies the MP3 files by Genre or category, such as
‘Rock’, ‘Jazz’, etc. Because of standard MP3 tags/terms and the MP3 genres or
classifications, it is possible to easily search for MP3 songs on the internet
or in a file system. Additionally, many software applications can read an MP3
library collection, import it, and automatically classify and sort MP3s based
on the metadata tags. Similarly, by using both a standardized set of metadata
terms for tagging and a set of published content classification categories,
health science content items (such as documents, images, photos, and other
digital media) can be tagged, organized, and more efficiently searched.
As in the MP3 content classification example, the OASIS eTMF
content model uses a published set of content classification categories in a
hierarchy, as shown in Figure 12. Each content classification primary category
contains Sub-Categories and Content Types. A Content Type is linked to a
reusable collection of metadata for a category of items or documents. Content
Data Properties, or Metadata, describe the document or content item’s
attributes.
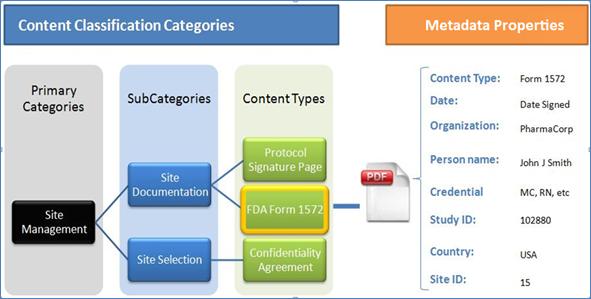
Figure 12: OASIS standard eTMF
Content Model Example
While the OASIS
eTMF content model hierarchy is flexible and interoperable, it is also useful
for organizations that want to save time and resources and share content models
with others.
The OASIS eTMF content models are created and
published as ontologies based on the W3C’s OWL 2.0 syntax and RDF/XML.
Semantic web allows seamless sharing, linking, and search of data across
domains. Additionally, the W3C’s OWL format is used in describing the content
model in order to retain compatibility with the technology that NCI, WHO, and
other leading health science vocabulary standards groups are using to model
information relationships (3) and (4) . The OWL format can be used to
represent hierarchies, complex relationships, data properties, and values. It
also allows search engine discovery and presentation in a web browser.
Furthermore, many Organizations are moving towards a semantic web model, which
enables interoperability between content models. For example, the CTMS model
follows BRIDG semantic web model; hence, the OASIS eTMF content model can be
more readily integrated with the CTMS for interoperability given they are both
based upon the semantic web model.
The best way to view, understand, and edit the OASIS eTMF
content model hierarchy is to open the content model in the Protégé OWL editing
application. Using the NCBO BioPortal/OASIS sites, download the OASIS eTMF
content model OWL file and open it in Protégé. In Protégé, users can add new
Categories, Content Type, and Data Properties, and perform editing operations
to change labels or other minor changes.
Protégé is a very sophisticated application. However, the OASIS
eTMF content model only uses a subset of the features of the OWL syntax and
Protégé. Users should focus primarily on the tabs ‘Classes’, ‘Individuals’, and
‘Data Properties’.
Figure 13 shows a screenshot of the Protégé editor. In this
example, we use the Stanford University’s Protégé application, freely available
for download at the following:
URL: http://protege.stanford.edu
[7].

Figure 13: Using Protégé OWL Editor to Modify OASIS eTMF
content Models
An example for a W3C RDF/XML file is illustrated in Figure 14. The W3C RDF/XML is used as the syntax for content model representation and
exchange. The file contains RDF and OWL in XML. Additionally, it includes
reference to content model Profile for the eTMF. Furthermore, the
RDF/XML file contains the content model instance for a study.
The .owl filename extension is used for the RDF/XML
files. Filenames for content model exchange shall be similar to IETF URL naming
as follows: Alphanumeric characters and the hyphen ‘-’ (special character) can
be used to ensure future compatibility.

Figure 14: Example W3C RDF/XML
File Snippet
The OASIS eTMF content model profile is represented as W3C OWL2 classes. In this way,
content models can be easily edited and shared by anyone. Content Model
instances are expressed as W3C
RDF/XML (e.g., eTMF specific study).
Generally, RDF/XML is used as the syntax for content model
exchange. Content models can be exchanged using Serialized RDF/XML or RDF/XML
as a file with .owl extension (e.g., etmf.owl). No specific exchange protocol
is specified by RDF/XML nor is one required for the content model exchange (the
protocol is application/implementation specific). Any protocol which supports
exchange of RDF/XML files or serialized data can be used (e.g., W3C http/s,
REST, SOAP, RPS, CMIS, etc.).
Versioning of content models is supported through the W3C
OWL Versioning
Policies . The W3C OWL supports granular level of versioning. However,
version management is considered to be an application-specific task. Being a
W3C standard, an OWL file includes an element for content model versioning; the
owl:versionInfo, which provides a hook suitable for use by version
management systems.
The OASIS content model version numbering text follows the
Major.Minor numbering format, where the Major part reflects the content model
profile version number. The Minor part should reflect an Organization-specific
version of the content model (as illustrated in Figure 15). This Minor
numbering may be enhanced with Organization-specific and application-specific
numbering within the W3C OWL versioning policies. The element owl:versionInfo
in RDF/XML should be used for Categories, Content Types, Annotation Properties,
and Metadata Propertiers. Finally, the first version of the OASIS eTMF content
model would be published as Version 1.0.
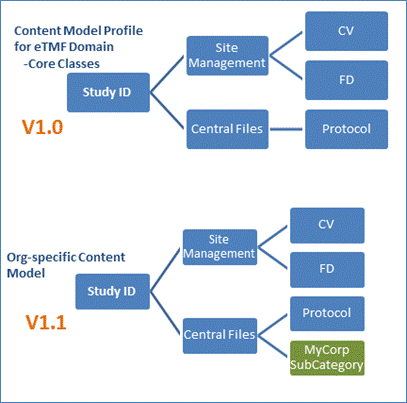
Figure 15: Content Model
Versioning Example
The Web Standard Technology Core Layer has three components:
the Data Model, the Electronic and Digital Signatures technologies, and
Business Process Model support.
The OASIS eTMF data model represents a single instance of an
eTMF content model for a single clinical trial. An eTMF data model instance
contains data values for metadata properties and content items for this
clinical trial study. It also includes the core and organization specific
content model categories and content types. The OASIS eTMF data model enables
organizations to package, archive, and share clinical trial records with other
systems or with regulatory agencies. For FDA part 11 compliance, the eTMF
electronic archives can be exported using common file formats of XML and PDF.
These electronic eTMF archives have the ability to be viewed in a simple web
browser or exchanged using simple files and folders that are operating system
and application independent. The information in all data models should contain
the content model instance, content item URIs and metadata values, and should
follow content classification rules and policies.
The OASIS eTMF content classification system uses W3C OWL2
RDF/XML to represent content models. Starting with standards based content
model ontology; it is possible to create an organization specific instance of
the content model that represents the clinical trial taxonomy, metadata values,
and content items in a clinical trial. As well as exporting records as
regulatory agency compliant archives, the OASIS eTMF clinical trial data model
can be exported to other systems that can be viewed in any web browser,
Exported OASIS eTMF data model content items can be stored in the cloud, simple
file folders, or other physical media such as network file shares or connected
storage with a URI.
The OASIS eTMF data model primary exchange file format is
the RDF/XML. The file includes core and organization specific Categories and
Content Types (reserved and in use), annotation properties, metadata properties,
and links to instance resource content items offline and online (linked data).
Content item name is unspecified and the content item file format is any
supported IANA media type format [1]. To allow exporting content items that use
the electronic Common Technical Document (eCTD) compatible file formats (for
FDA Part 11 compliance), a metadata term called eCTD Item tags eCTD
docs/records (tag value is true or false).
This format enables the interoperable exchange of content
models, content items from cloud or physical media, metadata terms, and
metadata values for clinical trial study instances between systems and
applications. Exported records are in XML format and no specific format for
content items is specified.
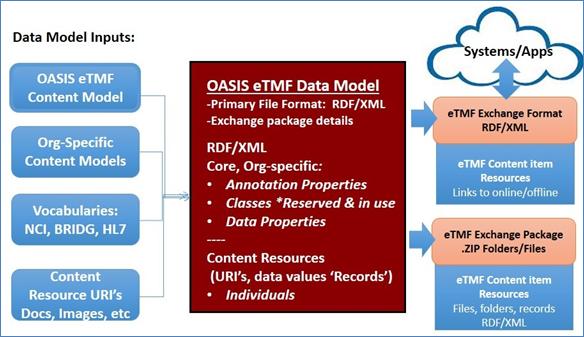
Figure 16: Data Model inputs, file format; eTMF Exchange
format and package
All OASIS eTMF content can be exchanged as collection of
folders following the folder taxonomy and includes content items and records in
RDF/XML format. Folders and resources are packaged in a standard .ZIP file,
with or without encryption. Additionally, the OASIS eTMF standard supports an Alternate
Taxonomy; alternate names for classification categories and content types
for the exchange format. The Alternate Taxonomy is supported through the use of
Display Name metadata property. This metadata property allows the use of
any language and names for the exported taxonomy. It can be used for any
category and content type for exchange. Interoperability is enforced with
RDF/XML. Figure 17 shows an example for the Alternate Taxonomy for the eTMF
Exchange Package folder names.
The eTMF exchange file format has a default structure and
naming and supports any IANA media type; thus, enabling broad flexibility. The
eTMF exchange file format will be an RDF/XML file with records and URI pointers
to linked content online or offline.

Figure 17: Alternative Taxonomy
Example
Electronic and digital signatures enable the removal of wet
signatures on paper. An electronic signature (i.e., point of sale signature) is
a digital mark accepted by many agencies. They are accepted by both EMA and
FDA. However, the signing party cannot be easily verified.
Digital signatures use a digital certificate issued to the
signing party and incorporate standards based technology (x.509 PKI). A digital
certificate is a cryptographic artifact. It contains a cryptographic key using
RSA or Elliptic Curve technologies, where the private key is not discernable by
knowing one’s public key. The subscriber gets a public key and a private key.
The digital certificate technology is known as the Public
Key Infrastructure (PKI). Currently, the FDA requires a digital signature for
eSubmissions and a trusted digital certificate on the documents themselves.
There are multiple types of digital certificates that are trustworthy. An
electronic document, such as a PDF, that is digitally signed using a digital
certificate from a recognized certificate authority is more reliable than a wet
signature on a paper. As of September 2013, the EMA moved to require only
digital certificates from recognized certificate authorities for three types of
submissions: orphan medicines, pediatric submissions, and scientific advice.
Digital certificates are issued to individuals, groups, and
devices. In the context of the OASIS eTMF standard content model, digital
certificates are issued to individuals to sign electronic documents. Digital
certificates must satisfy the EU qualified certificate policy and the EU
advanced electronic signature directive. As of the date of this document, there
is a complete overlap between these EU requirements and the FDA PKI policy.
Digital signatures enable validation of the signing party
through digital certificate technologies using a third party certificate
authority validation website. Contrary to electronic signatures, digital
signers are validated every time they sign. Additionally, digital signing
should use the two factor authentication, as per FDA CFR 21 Part 11.
The OASIS eTMF standard supports two types of electronic
signatures; electronic signatures (under Part 11 and EMA) and digital
signatures. The standard recognizes images with signatures, which can be
supported in the OASIS eTMF standard through metadata to indicate documents
that contain a scanned signature.
Generally, electronic signatures should comply with FDA and
EU regulations. Additionally, any file format approved by FDA and EU is
supported for e-signing. Digital signatures should use x.509 PKI certificates,
should comply with FDA and EU regulations, and should support any file format
approved by FDA and EU for e-signing. While not the purview of version 1.0 of
this draft standard, V2.0 of the OASIS eTMF standard should support EU
compliant Digital Signatures per emerging EU regulations.
Many organizations use automated business processes or
workflows for specific operations, such as electronic document approvals. The
Business Process Model component provides a mechanism to capture basic business
process model task completion information found in metadata linked to
documents. The Business Process Model is based on the Business Process Model
and Notation (BPMN) V2.0 specification, which is maintained by the Object
Management Group [7]. BPMN is a standard for business process modeling, i.e.,
the representation of processes in an enterprise, with an objective to support
business process management. The BPMN V2.0 includes two elements: Business
Process and Task. A Business Process is a collection of related tasks that
produce a specific output, e.g., a service or a product. A Task is a unit of
work that cannot be broken down into a further level of business process
details. An example is provided in Figure 18 to illustrate Business Processes
and Tasks. In this figure, the Business Process “Sign and Review” is split into
three Tasks: Send email request for Signature, Sign Document, and Review
Complete.
The aforementioned BPMN elements are supported in the OASIS
eTMF Business Process Model component. They are mapped to their equivalent OASIS
eTMF model Business Process Metadata (BPM) terms. For example, whenever a
process, or a specific task in the process, is completed for a particular
content item, a date/time stamped entry is captured in the OASIS eTMF model
BPM, forming an auditable document history log (see Figure 19 for an example).
The OASIS eTMF model Business Process component utilizes XML to capture the
details of BPMN processes and tasks. No specific software, system, or language
is required to implement it.
The OASIS eTMF model BPM enables capturing details about
business processes and tasks associated with a specific document. It also
allows organization and person entities to be associated with processes and
tasks. The OASIS eTMF model BPM includes Process Name, Task Name, Content Type,
Organization, Person, Source, Digital Signature, and Date. The OASIS eTMF model
BPM can be captured for any Content Type.
When used in conjunction with digital signatures, the OASIS
eTMF model Business Process Component offers automation of paper-based document
approval and signing processes. Figure 18 shows an example of a BPMN V2.0
process for an automated signature approval on a confidentiality agreement. Figure 19 illustrates how the BPMN signature process example in Figure 18 is mapped into BPM, with a date/time stamp captured to indicate completion of each task. By
capturing the date/time stamp in the BPM for each completed process and task,
each document resource has its own auditable workflow metadata history; this
enables detailed auditing and reporting in applications. For each Content Type
that uses BPM, a new entry is made in the BPM history log for each task in a
process. For example, one of the tasks is Send email request for signature,
which is associated with the Content Type ‘Confidentiality Agreement’. This
information is captured in the OASIS eTMF model BPM history log as shown in the
figure.
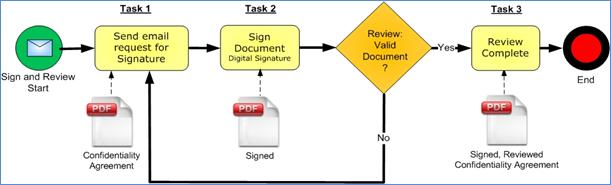
Figure 18: Business Process Management “Sign and Review Process” Example Using the BPMN
Notation
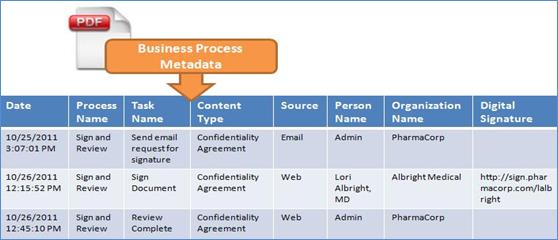
Figure 19: The OASIS eTMF BPM
Audit Trail Log Captured for Tasks in Figure 18
An implementation is a conforming eTMF Content
Classification System if the implementation meets the conditions in section
1.1:
1.1 Conformance as an
eTMF Content Classification System
An implementation is a conforming eTMF Content
Classification System if the implementation meets the conditions:
a) Conforms to
specifications detailed in the OASIS eTMF Content Classification System
(CCS) Layer, with support for the Content Classification System, the
RDF/XML based Content Model and includes support for the Core Metadata.
I.
Conforms
to CCS specifications for naming, numbering and organizing content
classifications
II.
Conforms
to CCS specifications and policy rules for modifying and editing content
classification entities.
III.
Includes
Core Metadata as property tags for all content classified in the system.
b) Conforms to
specifications detailed in the OASIS eTMF Metadata Vocabulary Interoperability
Layer, and sources core classification terms from the published OASIS eTMF
Standard Ontology, based on terms published at the National Cancer Institute’s
NCI Thesaurus term repository.
I.
Supports
the Metadata Vocabulary Interoperability Layer requirement that
classification entity terms must have a user-modifiable display label to enable
localized terms to end-users.
c) Conforms to
specifications detailed in the OASIS eTMF Web Standard Core Layer.
I.
Supports
content exchange through the RDF/XML based data model component.
II.
Supports
FDA and EMA acceptable electronic and digital signatures as detailed in the
electronic and digital signature component.
d) If the eTMF Content
Classification System is used for any application where ‘individually
identifiable health information’ is captured, stored or transmitted, US
Department of Health and Human Services HIPAA privacy regulations and
compliance policies for electronic records and patient protected health
information (PHI) security must be implemented in the eTMF System. For
example, investigator site data may contain individually identifiable patient protected
health information. System protections and data management policies for the
protection of this information must be implemented to be in compliance with US
HIPAA privacy regulations.
e) If the eTMF Content
Classification System is used for clinical trials subject to FDA regulation and
electronic records and / or electronic signatures are used, the system must
adhere to applicable FDA regulations such as 21 CFR Part 11 or other related
agency regulations.
The following individuals have participated in the creation
of this specification and are gratefully acknowledged:
|
Participant:
|
Organization:
|
|
Sharon
Ames
|
NextDocs
|
|
Michael
Agard
|
Paragon
Solutions
|
|
Jennifer
Alpert Palchak
|
CareLex
|
|
Peter
Alterman, PhD
|
SAFE-BioPharma
Association
|
|
Aliaa
Badr
|
CareLex
|
|
Lou
Chappuie
|
SureClinical
|
|
Sharon
Elcombe
|
Mayo
Clinic
|
|
Chet
Ensign
|
OASIS
|
|
Robert
Gehrke
|
Mayo
Clinic
|
|
Troy
Jacobson
|
Forte
Research Systems, Inc.
|
|
Rich
Lustig
|
Oracle
|
|
Christopher
McSpiritt
|
Paragon
Solutions
|
|
Jamie
O'Keefe
|
Paragon
Solutions
|
|
Oleksiy
(Alex) Palinkash
|
CareLex
|
|
Fran
Ross
|
Paragon
Solutions
|
|
Catherine
Schmidt
|
SterlingBio
|
|
Zack
Schmidt
|
SureClinical
|
|
Mead
Walker
|
Health
Level Seven, Inc.
|
|
Trish
Whetzel, PhD
|
SureClinical
|
The Technical Committee thanks the
following for their work:
For its work on the eTMF controlled vocabulary,
we acknowledge the
National Cancer Institute, Enterprise Vocabulary Services. In particular, we acknowledge the work
of:
Margaret W. Haber, Program Manager,
Enterprise Vocabulary Services, National Cancer Institute
Theresa Quinn, Biomedical/Clinical
Research Information Specialist (C) Enterprise Vocabulary Services, National
Cancer Institute
Jordan Li, PhD, Biomedical/Clinical Research
Information Specialist (C) Enterprise Vocabulary Services, National Cancer
Institute
Erin Muhlbradt, PhD,
Biomedical/Clinical Research Information Specialist (C) Enterprise Vocabulary
Services, National Cancer Institute
For its work in reviewing the specification, we acknowledge
the PhUSE/FDA
project. In particular, we
acknowledge the work of Kerstin L. Forsberg
B.1 OASIS eTMF Classification Terms
Classification Category terms, used in the OASIS eTMF
Content Model, are sourced from NCI (1) using the CareLex Preferred Term, as
approved by the eTMF Technical Committee. Classification terms are published
online and curated by NCI.
B.2 OASIS eTMF Model Metadata Properties
The OASIS model Metadata is linked to each Content Type.
Similar to how MP3 files are tagged with metadata like artist, song, etc., the
purpose of the OASIS model Metadata is to tag content items for use in
classification, navigation, searching, and reporting. The OASIS model Metadata
is split into Core Metadata, General Metadata, Domain-specific, and
Organization-specific Metadata. The OASIS Metadata types and their uses are
illustrated in Table 7. In the following, Core Metadata is defined in Table 8, eTMF Metadata (i.e., domain-specific metadata) is represented in Table 9, and General Metadata is illustrated in Table 10.
Table
7: OASIS Model Metadata Types and Usage
|
Metadata Type
|
Description
|
How Used
|
|
Core
|
Core metadata are split into four main areas as follows:
· File Properties:
The metadata present in most digital files, such as Created, Modified,
Filename, Format, and URI (i.e., path).
· Basic audit trail:
Indicates username for the user who created and modified a content item.
Fields are Created by and Modified by. These are widely used fields in
popular CMS’s data properties.
· Classification:
Terms used in classification, e.g., Category and Sub-Category. These are
annotation properties. When used with a content model, it enables automated
content classification and metadata assignment for an item’s Annotation
Properties.
·
Business
Processes: A set of metadata properties that enables capturing tasks in business
or workflow processes for a Content Type. For example, after a task (like
submittal of an approval or a signature) is completed for a content item, an
entry is made in the business process metadata history log to record the
task’s occurrence. Additionally, each task includes a date stamp and the name
of the organization and/or person to whom the task is linked. For example, in
a digital signing business process, a single business process metadata
history log entry can be completed using the following attributes:
§ Date: Date of
signing event.
§ Task: The task
associated with the document, such as Signed, Approved, Declined, etc.
§ Source: The source
of the content item, such as Import Scan, Fax, E-mail, System, and Other.
§ Organization Name:
The name of the organization which performed the business process task or to
whom the item or document is linked. Note that Organization name should be
captured for each document.
§ Person Name: The
name of the person who performed the business process task to whom the document
is linked.
§ Digital Signature:
The optional URI to digital certificate information for the document.
Each document must contain at
least a Date and Organization Name to which the item is linked. Additionally,
each event creates a workflow history entry, in the workflow history table,
which is captured for each content item.
|
Use differs according to the area:
· File Properties:
Same as in digital files. It enables minimal metadata mapping for operations,
such as drag and drop of files.
· Basic audit trail:
Same as the popular EDMSs’ basic audit information for an item. They enable
minimal metadata mapping for operations, such as drag and drop of files.
· Classification:
Used in automated content classification.
·
Business
Process: Used in the following:
§ Content
Item business process or workflow history.
§ Content
item enhanced audit trail.
§ Content
Item digital signing process.
|
|
Domain-specific
|
Metadata
used in specific domain areas, such as the metadata used in the Clinical
Trials electronic Trial Master File (eTMF) of the health science domain. This
type of metadata varies according to the domain area.
|
It provides domain specific information for content items,
e.g., Clinical Study Report metadata (used in the eTMF content model in the
health science domain) describes final or interim results of a trial.
|
|
General
|
The set of
metadata properties obtained from public terminology resources and metadata
vocabularies (in support of interoperable solutions), such as Dublin Core
Metadata. This type of metadata properties can be applied to a content type
at discretion of the implementer.
|
It provides metadata that can be used in any Content
Model. As an example, the Dublin Core resource provides “Description”
metadata, which supplies an account for the content item.
|
|
Organization-specific
|
User
defined metadata that enables organizations to define their own metadata to
fulfill their needs.
|
|
B.2.1 Core Metadata
The following Table illustrates Core Metadata found in all
content models irrespective of domain area.
Table 8: Core Metadata
|
Term
|
Definition
|
Code
|
|
File Properties
|
|
Created
|
The date and time at which the resource is created. For a
digital file, this need not match a file-system creation time. For a freshly
created resource, it should be close to that time. Later file transfer,
copying, etc., may make the file-system time arbitrarily different.
|
C69199
|
|
Modified
|
The date and time the resource was last modified.
|
C25446
|
|
Content Identifier
|
The unique identifier for a content item, such as a
document, image, or other media in a specified context. (Document name.)
|
C99023
|
|
URI
|
The unique uniform resource Identifier or path (URI) for a
content item such as a document, image, or other media in a specified
context.
|
C42778
|
|
Format
|
Content Item File Format, e.g., PDF, JPG, GIF, XLS, DOC,
DOCX, XLSX, PPT, PPTX. It uses a filename extension as the format value.
|
C42761
|
|
Document
Version
|
A
representation of a particular edition or snapshot of a document as it exists
at a particular point in time. Please refer to the Appendix for details
regarding the document versioning policies.
|
C93484
|
|
Country Code
|
Name of country using ISO 3166-1 alpha-3 country codes-
Example: USA.
|
C20108
|
|
Basic Audit
Trail
|
|
Created By
|
Indicates the username of the person who brought the item
into existence.
|
C42628
|
|
Modified By
|
Indicates the username of the person who changed an item.
|
C42629
|
|
Classification
|
|
Content Type Name
|
The name of the Content Type such as 'CV.' A Content Type
is a reusable collection of metadata, workflow, behavior, and other settings
for a category of items in electronic content material.
|
C115999
|
|
Business Process
|
|
Date
|
Date of
task or event, or date in the context of document or Content Type. Date can
be different from date created.
|
C25164
|
|
Process
|
A sequence
or flow of activities in an organization with the objective of carrying out
work. Source: BPMN V2.0 Spec (4). Tasks are atomic activities. They are
included within a Process.
|
C29862
|
|
Task
|
A single
activity that has occurred within a business process. Generally, an
end-user, an application, or both will perform the Task. Concept derives
from BPMN V2.0. Example task values are: Submitted, Approved, Reviewed,
Signed, etc., indicating that a task has been completed. Each task is date
stamped and captured in a single record of the business process metadata
history log.
|
C101129
|
|
Source
|
Where the
content item is from or its origin. Example values: Import, Scan, Fax,
email, system, and other.
|
C25683
|
|
Person
Name
|
The full
name of the person who performed the workflow action (e.g., approved or
submitted a document) or the person to whom this document is linked.
|
C25191
|
|
Person
Role
|
The role
of the person who is responsible for or linked to a content item, such as
Principal Investigator, Sub-Investigator, Study Coordinator, Sponsor Project
Manager, CRO Project Manager, or Data Manager.
|
C113644
|
|
Subject
Identifier
|
Subject
Identifier is a unique sequence of characters used to identify, name, or
characterize the study subject individual in a clinical trial study.
|
C83083
|
|
Organization
Name
|
The full
name of the Organization linked to the resource.
|
C93874
|
|
Organization
Role
|
Denotes
the role of the organization, which is responsible for or linked to the
Content Item. Values include Sponsor, Site, CRO, and Vendor.
|
C114551
|
|
Username
|
The
account name used by a person to access a computer system (used for system
generated tasks).
|
C42694
|
|
Digital
Signature
|
Extra data
embedded in a document or metadata linked to a document. It identifies and
authenticates the signer of a document using public-key encryption. May be a
URI or path to digital signature resource or certificate.
|
C80447
|
|
Digital
Signature Status
|
Specifies
whether a document or content item has been digitally signed. If no
signature is required, status = null. Values: Signed, Not Signed, Null
|
C114552
|
|
|
|
|
B.2.2 Domain based Metadata (eTMF Domain Example)
Note that the OASIS eTMF model can support other domains in
life sciences or healthcare such as research and development, finance,
administration, or others.
Table 9: Metadata Used in the eTMF Domain Area (Content Model = eTMF)
|
Term
|
Definition
|
Code
|
|
Study ID
|
A sequence of characters used to identify, name, or
characterize the study.
|
C83082
|
|
Site ID
|
A unique symbol that establishes the identity of the study
site.
|
C83081
|
|
Credential
|
Professional
credential of Person for study - MD, RN, PhD or other for Person linked to a
content item / document; EX: MD, RN, PhD, MS, MA, BA, MBA
|
C73925
|
|
Visit
Number
|
The
numerical identifier of the visit.
|
C83101
|
|
Clinical Study Sponsor
|
An entity that is responsible for the initiation,
management, and/or financing of a clinical study
|
C70793
|
|
eCTD Item
|
An electronic content item and its associated electronic
record that is targeted for inclusion in an Electronic Common Technical
Document. Valid values are Yes/No..
|
X90010
|
B.2.3 General Metadata
This section discusses General Metadata (obtained from
public terminology resources and metadata vocabularies) that can be used in OASIS
eTMF ontologies. Terms presented in Table 10 are from the Dublin Core Project [4].
Table
10: General Metadata in Dublin Core
|
Term
|
Definition
|
Code
|
|
Description
|
An account of the resource or content item.
|
X90005
|
|
Location
|
A spatial region or named place.
|
X90006
|
|
Title
|
A name given to the resource or content item.
|
X90007
|
|
Type
|
The nature or genre of the resource or content item.
|
X90008
|
B.3 Annotation Properties
Table 11: Annotation
Properties
|
Term
|
Definition
|
Code
|
|
Abbreviation
|
Preferred Abbreviation of term name.
|
C42610
|
|
Annotation Property
|
Annotation Properties are used to provide annotations for
a content model entity or term. For more, refer to general description at:
http://www.w3.org/TR/owl2-syntax/#Annotation_Properties
|
C114458
|
|
Archive
|
A single collection of related digital content items
within a digital repository, for example a collection of clinical trial
documents for a single clinical trial Study. File based or database. Child
entities are Categories, Organization, Person, Data Property, and Annotation
Property.
|
C114463
|
|
Category
|
A hierarchical classification of Content Items within an
Archive, with child Sub-Categories and Content Types. Child entities =
Sub-Category, Data Property assignments, Data Property values, Annotation
Property assignments, and Annotation Property values.
|
C25372
|
|
Category Code
|
A coded value specifying a Category or Content Type
classification using the OASIS eTMF Standard Classification numbering
scheme. Required for every Category and Content Type, e.g., 100.11.
|
C93524
|
|
Category Name
|
The word or phrase used by preference to refer to an
entity or a term, including Category, Content Type, Data Property, Annotation
Property, Organization, and Person.
|
C42614
|
|
Category Type
|
Represents the type of the classification Category, which
could either be Core (i.e., part of the published Content Model),
Domain-specific (i.e., added to the Content Model from another model), or
Organization-specific (i.e., user defined). Possible values are core, domain,
and org.
|
C115618
|
|
Code
|
Unique Code assigned to each term, including Category,
Content Type, Data Property, or Annotation Property. NCI Thesaurus codes
begin with ‘C’; CareLex term codes begin with X; Codes which are pending
review by CareLex begin with ‘Y’, while Organization-specific codes and codes
for terms used as General metadata (those with no previously assigned Code)
begin with ‘Z’. All codes are 1 alpha char (a-z) and 5 numeric characters
0-9.
|
C115478
|
|
Content Item
|
A single digital file, which may include digital
documents, images, multimedia, or other digital content. It has one and only
one Content Type parent. Child entities = Data Property assignments and Data
Property values.
|
C99095
|
|
Content Model
|
Specifies the content model that the term belongs to. A
content model defines how content can be classified for a specific business
domain. It is similar to an electronic filing system folder structure, where
content or documents are classified and assigned properties. A content model
is structured as ontology comprised of a hierarchical taxonomy of content
categories, content or document types, and data properties or metadata. The
relationships between content types and data properties are described in a
machine readable computer language known as RDF/XML.
|
C114459
|
|
Content Type
|
A content type is a reusable collection of metadata,
business processes, behavior, and other settings for a category of items or
documents in electronic content material.
|
C114466
|
|
Creator
|
Specifies the name of a person or entity that enters a
term in the content model ontology.
|
C42628
|
|
Cross Reference
|
Optional Code indicating mapping of term to an external
term in another place (if more than one, each code separated by semi-colon).
|
C43621
|
|
Data Property
|
Data Properties are similar to XML attributes. They are
used to define the attributes of Content Types. For more information, refer
to the general description at: http://www.w3.org/TR/owl2-syntax/#Data_Properties
|
C114468
|
|
Data Type
|
A data type is a classification identifying one of
various types of data, such as string, date, integer, or Boolean. It
determines the possible values for that type of value.
|
C42645
|
|
Definition
|
A concise explanation of the meaning of the Data Property.
|
C42777
|
|
Display Name
|
Provides an alternative Preferred Name. For use by
software or systems to provide a localized display name. By default, Display
Name is the same as Preferred Name.
|
C70896
|
|
Label
|
Used only in OWL RDF/XML file. An identifying marker
attached to a Category, Content Type, Data Property or Annotation Property.
Label is always the same as Preferred Name and cannot be changed.
|
C45561
|
|
Metadata
|
Used in Ontology to indicate which metadata or data
property attributes are assigned to a given content type. Metadata
attributes are assigned to Content Types in RDF/XML separated by semi-colons.
|
C52095
|
|
Metadata Type
|
Metadata terms are organized by type. Types of metadata
are: Core metadata, Business Process Metadata, Domain Specific metadata,
General Metadata, Organization Specific.
|
C114465
|
|
Preferred Name
|
The word or phrase used by preference to refer to an
entity or a term, including Category, Content Type, Data Property, Annotation
Property, Organization, and Person.
|
C43707
|
|
Regulation
|
Specifies agency regulatory Code Section that corresponds
to a Content Type requirement for example, FDA CFR 21 part 11, ICH 5.1.1.
Multiple Code sections are separated by semi-colon characters.
|
C68821
|
|
Repository
|
A digital library in which collections, or archives, of
content items are stored in digital format. File based or database. Child
entities = Archive.
|
C114457
|
|
Requirement
|
For a Content Type, specifies whether the Content Type is
required by a regulatory agency or to satisfy organization requirements.
Does not apply where content type = category. Valid values: Required,
Optional, Required if Applicable
|
C115762
|
|
Sub-Category
|
A classification of content within a parent Category. It
may have zero or more Content Types as children. Additionally, it has zero or
more Sub-Categories. It is the child of Category.
|
C25692
|
|
Synonym
|
Synonyms for the Category, Content Type, Data Property or
Annotation Property
|
C52469
|
|
Term Source
|
An indicator of the particular group or agency that
supplied a specific term.
|
C43822
|
|
Term Source URL
|
Indicates URL where term is defined and maintained.
Organization specific terms do not require Term Source URL.
|
C114456
|
|
Term Type
|
The type of term. Valid values are Primary Category,
Sub-Category, Content Type, Data Property, Annotation Property, and Entity.
|
C114455
|
|
Value Set
|
A value set is a fixed set of data values that can be
selected for a given data property attribute. The value set helps ensure
data entry accuracy for a given data property field. For example, the data
property Organization Role may have a value set of: sponsor, investigator,
CRO. The data property attribute could be assigned one of these value set
values.
|
C113497
|
B.4 Document Version Numbering Policies
In the OASIS eTMF Standard, the document version text values
follow the same formatting that is familiar and commonly implemented in
software and in other health science standards: Major Version.Minor Version.
Version numbering text is an integer value separated by a period, without
leading zeros. There can be a new Major version every time the
document/content item changes. There can be a new Minor version every
time the metadata changes.
Within the OASIS eTMF archives, content item version
management shall be application specific to provide application flexibility.
However, for consistent content item exchange, version number text formatting
should be implemented using the OASIS eTMF document version numbering policies
(based on NCI/CDISC/FDA/BRIDG definition: C93816):
- Each document Major version number
is an integer starting at '1' and is incremented by 1. The first
instance, or the original content item, should always be valued as '1'.
The version number value must be incremented by one when a document is
replaced, but can also be incremented more often to meet application
specific requirements.
- Different versions of the same
document belong to the same Content Type group.
- The document Minor version number
would be an integer starting at ‘0' and incrementing by 1. The first
instance of an original document with no minor version should always be
valued as ‘1.0’, where ‘0‘indicates that no minor version exists.
- Documents with a change to the metadata
values would require a minor version. The first minor version for a
1.0 document would be indicated as 1.1. Successive changes to any of the
document’s metadata would increment the Minor version by 1. For example
1.2 indicates major version 1 and minor version 2. The Minor version
number value must be incremented by one when a document’s metadata is
changed, but can also be incremented more often to meet the application
specific requirements.
|
Term
|
Definition
|
Reference
|
|
EDMS
|
Electronic Document Management System: a collection of
technologies that work together to provide a comprehensive solution for
managing the creation, capture, indexing, storage, retrieval, and disposition
of records and information assets of the organization.
|
http://www.nd.gov/itd/services/enterprise-document-management-system-edms
|
|
eTMF
|
Electronic Trial Master File: is a formalized means of
organizing and storing documents, images, and other digital content for
pharmaceutical clinical trials that may be required for compliance with
government regulatory agencies.
|
http://en.wikipedia.org/wiki/Electronic_trial_master_file
|
|
eCTD
|
Electronic Common Technical Document: is CDER/CBER’s standard
format for FDA approved electronic regulatory submissions.
|
http://www.fda.gov/Drugs/DevelopmentApprovalProcess/FormsSubmissionRequirements/ElectronicSubmissions/ucm330116.htm
|
|
NCI
|
National Cancer Institute: is part of the National
Institute of Health (NIH) and coordinates the U.S national Cancer program and
conducts and supports research, training, health information dissemination,
and other activities related to the causes, prevention, diagnosis, and
treatment of Cancer.
|
http://en.wikipedia.org/wiki/National_Cancer_Institute
|
|
NCI EVS
|
NCI Enterprise Vocabulary Service: provides terminology
content, tools, and services to code, analyze, and share cancer and
biomedical research, clinical, and public health information.
|
http://evs.nci.nih.gov/
|
|
Revision
|
Date
|
Editor
|
Changes Made
|
|
R01
|
April 18 2014
|
Aliaa Badr
|
Working draft – first version
|
|
R02
|
May 24 2014
|
Aliaa Badr
|
Working draft – apply edits by Jennifer, Rich, and Airat.
|
|
R03
|
June 7 2014
|
Aliaa Badr
|
Updated graphics (Figure 6 and 7) to reflect correct
classification terms. Added sub-section in the Appendix for OASIS
Classification Terms
|
|
201406-R01
|
June 13, 2014
|
Zack Schmidt
|
Renamed document and version for public review as
committee specification draft
|





















